planarian = sc.read_h5ad(os.environ["EXAMPLE_DATA_PATH"] + "planarian.h5ad")
planarian.var.index = "pl_" + planarian.var.indexplot
See the big picture
These functions are intended to help you find your way around the data. I use them a lot when generating reports for collaborators, to set the stage for the more detailed analyses. In particular, I want to highlight which clusters we are comparing and where they are in the context of both species as well as the comparison.
highlighted_dimplot
highlighted_dimplot (adata:anndata._core.anndata.AnnData, species:str, clustering:str, cluster:str, embedding:str='X_umap', highlight:str='red', figsize:tuple=(10, 10), save:Optional[str]=None)
Plot a low-dimensional embedding and highlight a chosen cluster with a superimposed circle.
| Type | Default | Details | |
|---|---|---|---|
| adata | AnnData | AnnData object to plot. | |
| species | str | Species name. Will be used in the title, and removed from the cluster names if present. | |
| clustering | str | Clustering to plot. Must be present in adata.obs. |
|
| cluster | str | Cluster to highlight. | |
| embedding | str | X_umap | Embedding to plot (default: “X_umap”). |
| highlight | str | red | Color of the circle (default: “red”). |
| figsize | tuple | (10, 10) | Figure size (default: (10, 10)). |
| save | typing.Optional[str] | None | Path to save the figure (default: None). |
| Returns | None |
highlighted_dimplot(planarian, "Smed", "tissue", "Pharynx", figsize=(6, 6))/opt/homebrew/Caskroom/mambaforge/base/envs/ascc24/lib/python3.9/site-packages/scanpy/plotting/_tools/scatterplots.py:1234: FutureWarning: The default value of 'ignore' for the `na_action` parameter in pandas.Categorical.map is deprecated and will be changed to 'None' in a future version. Please set na_action to the desired value to avoid seeing this warning
color_vector = pd.Categorical(values.map(color_map))
/opt/homebrew/Caskroom/mambaforge/base/envs/ascc24/lib/python3.9/site-packages/scanpy/plotting/_tools/scatterplots.py:394: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
cax = scatter(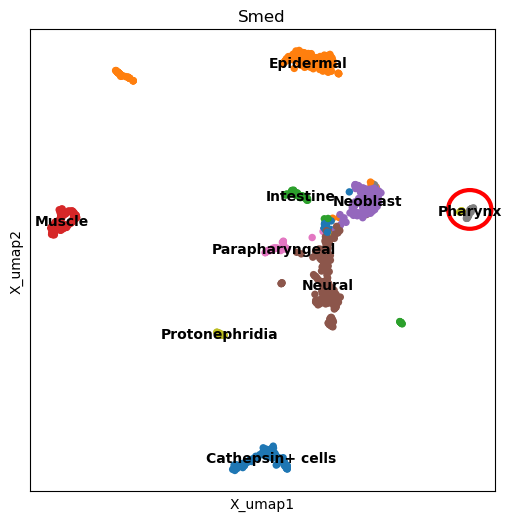
Of course, if a cluster is too spread out, the plot will not work as well, but it will still be useful to see the general location of the cluster.
highlighted_dimplot(planarian, "Smed", "tissue", "Neural", figsize=(6, 6))/opt/homebrew/Caskroom/mambaforge/base/envs/ascc24/lib/python3.9/site-packages/scanpy/plotting/_tools/scatterplots.py:1234: FutureWarning: The default value of 'ignore' for the `na_action` parameter in pandas.Categorical.map is deprecated and will be changed to 'None' in a future version. Please set na_action to the desired value to avoid seeing this warning
color_vector = pd.Categorical(values.map(color_map))
/opt/homebrew/Caskroom/mambaforge/base/envs/ascc24/lib/python3.9/site-packages/scanpy/plotting/_tools/scatterplots.py:394: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
cax = scatter(
Another case that might not work so well is if the shape of a cluster is not Gaussian. Still, it should help us spot the cluster, especially if the color contrasts enough.
highlighted_dimplot(
planarian, "Smed", "tissue", "Muscle", figsize=(6, 6), highlight="lightgreen"
)/opt/homebrew/Caskroom/mambaforge/base/envs/ascc24/lib/python3.9/site-packages/scanpy/plotting/_tools/scatterplots.py:1234: FutureWarning: The default value of 'ignore' for the `na_action` parameter in pandas.Categorical.map is deprecated and will be changed to 'None' in a future version. Please set na_action to the desired value to avoid seeing this warning
color_vector = pd.Categorical(values.map(color_map))
/opt/homebrew/Caskroom/mambaforge/base/envs/ascc24/lib/python3.9/site-packages/scanpy/plotting/_tools/scatterplots.py:394: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
cax = scatter(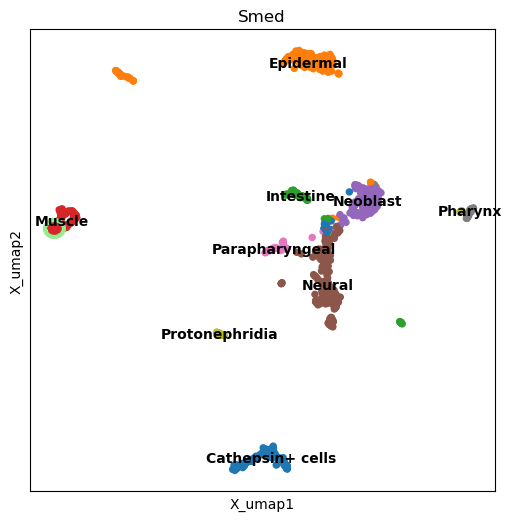
highlighted_heatmap
highlighted_heatmap (to_plot, celltype_from, celltype_to, figheight=20, save=None)
Plot a heatmap of pairwise similarities between cell types, with a red box highlighting the query cell type.
| Type | Default | Details | |
|---|---|---|---|
| to_plot | pd.DataFrame | A dataframe of pairwise similarities between cell types. | |
| celltype_from | str | Cell type of the query species to highlight. Must be in to_plot.columns. |
|
| celltype_to | str | Cell type of the target species to highlight. Must be in to_plot.index. |
|
| figheight | int | 20 | Height of the resulting plot in inches. Width will be calculated automatically (default: 20). |
| save | NoneType | None | Path to result figure; if None, the figure will be plotted but not saved (default: None). |
| Returns | None |
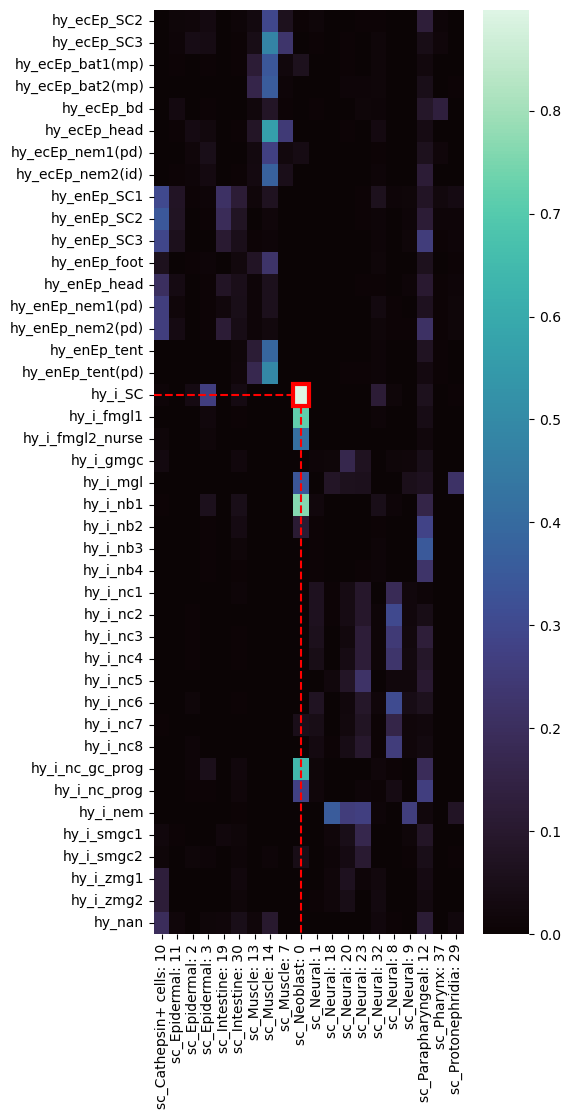
hysc = pd.read_csv(
os.environ["EXAMPLE_DATA_PATH"] + "hysc_similarity_table.csv",
index_col=0,
)highlighted_heatmap(
hysc, celltype_from="hy_i_SC", celltype_to="sc_Neoblast: 0", figheight=12
)
annotated_heatmap
annotated_heatmap (sm:samap.mapping.SAMAP, similarity:pandas.core.frame.DataFrame, query_species:str, target_species:str, query_clustering:str, target_clustering:str, query_coarse:Optional[str]=None, target_coarse:Optional[str]=None, interactive:bool=False, figsize:Optional[Tuple[float,float]]=None, save:Optional[str]=None, **kwargs:Any)
Plot the similarity matrix as an annotated heatmap.
| Type | Default | Details | |
|---|---|---|---|
| sm | SAMAP | SAMAP object | |
| similarity | DataFrame | Similarity matrix. Contains query species clusters as columns and target species clusters as rows. | |
| query_species | str | Query species ID. Will be used in the title. Should prepend the similarity matrix column names. | |
| target_species | str | Target species ID. Will be used in the title. Should prepend the similarity matrix row names. | |
| query_clustering | str | Query species clustering. Must be present in sm.sams[query_species].adata.obs. |
|
| target_clustering | str | Target species clustering. Must be present in sm.sams[target_species].adata.obs. |
|
| query_coarse | typing.Optional[str] | None | |
| target_coarse | typing.Optional[str] | None | Query species coarse clustering. Must be present in sm.sams[query_species].adata.obs. If None, will be set to query_clustering (default: None). |
| interactive | bool | False | If True, will return a plotly figure. Otherwise, will return a matplotlib figure (default: False). |
| figsize | typing.Optional[typing.Tuple[float, float]] | None | |
| save | typing.Optional[str] | None | Figure size. If None, will be guessed from the size of the similarity matrix (default: None). |
| kwargs | typing.Any | ||
| Returns | typing.Optional[plotly.graph_objs._figure.Figure] | If not None, will save the figure to the specified path (default: None). Additional arguments to pass to seaborn.heatmap (matplotlib) or plotly.graph_objects.Figure (plotly). Among them: dpi (int), which is only used if interactive=True to set the figure size in pixels. |
Read the requisite files: the SAMap object and the pairwise cluster similarity matrix.
file = open(os.environ["EXAMPLE_DATA_PATH"] + "hypl.pkl", "rb")
sm = pickle.load(file)
hypl = pd.read_csv(
os.environ["EXAMPLE_DATA_PATH"] + "hypl_similarity_table.csv",
index_col=0,
)First improve the coarse level assignments for the data we have here:
sm.sams["hy"].adata.obs["coarse"] = (
sm.sams["hy"].adata.obs["Cluster"].str.split("_").str[0]
)
sm.sams["pl"].adata.obs["coarse"] = (
sm.sams["pl"].adata.obs["tissue_smedwi"].str.split("_").str[0]
)annotated_heatmap(
sm,
hypl,
"hy",
"pl",
query_clustering="Cluster",
target_clustering="cluster",
query_coarse="coarse",
target_coarse="coarse",
interactive=False,
save="hypl.pdf",
)/opt/homebrew/Caskroom/mambaforge/base/envs/ascc24/lib/python3.9/site-packages/scanpy/plotting/_tools/scatterplots.py:1234: FutureWarning: The default value of 'ignore' for the `na_action` parameter in pandas.Categorical.map is deprecated and will be changed to 'None' in a future version. Please set na_action to the desired value to avoid seeing this warning
color_vector = pd.Categorical(values.map(color_map))
/opt/homebrew/Caskroom/mambaforge/base/envs/ascc24/lib/python3.9/site-packages/scanpy/plotting/_tools/scatterplots.py:394: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
cax = scatter(
/opt/homebrew/Caskroom/mambaforge/base/envs/ascc24/lib/python3.9/site-packages/scanpy/plotting/_tools/scatterplots.py:1234: FutureWarning: The default value of 'ignore' for the `na_action` parameter in pandas.Categorical.map is deprecated and will be changed to 'None' in a future version. Please set na_action to the desired value to avoid seeing this warning
color_vector = pd.Categorical(values.map(color_map))
/opt/homebrew/Caskroom/mambaforge/base/envs/ascc24/lib/python3.9/site-packages/scanpy/plotting/_tools/scatterplots.py:394: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
cax = scatter(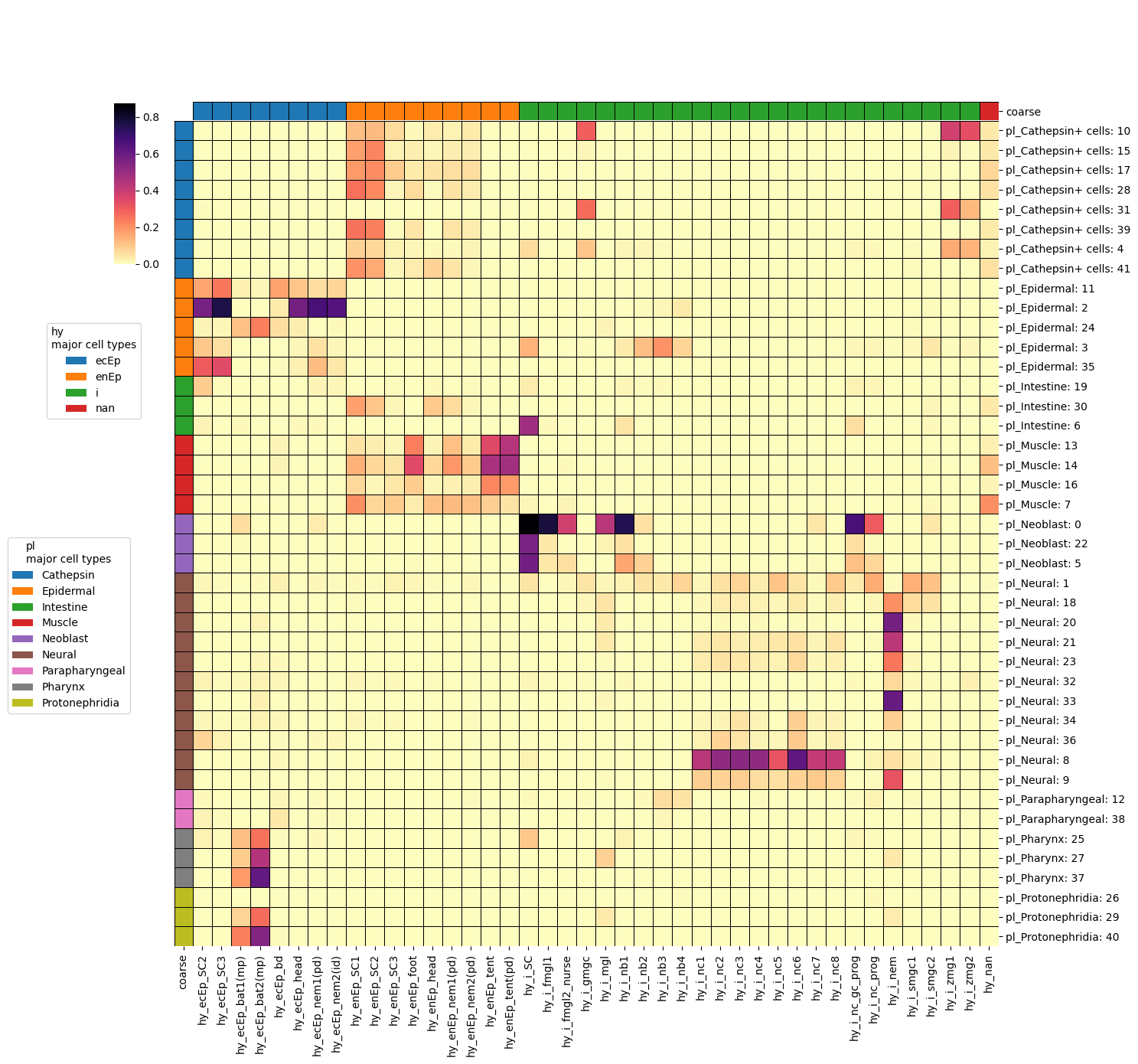
This sort of plot does a much better job of showing the relationships between clusters than the Sankey diagram. Clear patches of similarity emerge that would be otherwise invisible. This is a result of the hierarchical relationship between cell types (see Arendt/Musser/Wagner in 2016 and 2019). By combining maps like this with the per-species cell type family trees we can start to get a better idea of how cell types may have evolved over time.
assert os.path.exists("hypl.pdf")We can make the same plot in interactive form. Unfortunately, the interactive version is a bit more finicky with color usage, so for now I’ll let plotly choose the colors, but any plotly wizards out there are welcome to improve this. Right now hovering over a point will show the cluster names and SAMap score; hovering over the colored bars at the margins should show the coarse cluster.
plotly_overview = annotated_heatmap(
sm,
hypl,
"hy",
"pl",
query_clustering="Cluster",
target_clustering="cluster",
query_coarse="coarse",
target_coarse="coarse",
figsize=(10, 10),
interactive=True,
dpi=100,
save="hypl",
)/opt/homebrew/Caskroom/mambaforge/base/envs/ascc24/lib/python3.9/site-packages/scanpy/plotting/_tools/scatterplots.py:1234: FutureWarning: The default value of 'ignore' for the `na_action` parameter in pandas.Categorical.map is deprecated and will be changed to 'None' in a future version. Please set na_action to the desired value to avoid seeing this warning
color_vector = pd.Categorical(values.map(color_map))
/opt/homebrew/Caskroom/mambaforge/base/envs/ascc24/lib/python3.9/site-packages/scanpy/plotting/_tools/scatterplots.py:394: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
cax = scatter(
/opt/homebrew/Caskroom/mambaforge/base/envs/ascc24/lib/python3.9/site-packages/scanpy/plotting/_tools/scatterplots.py:1234: FutureWarning: The default value of 'ignore' for the `na_action` parameter in pandas.Categorical.map is deprecated and will be changed to 'None' in a future version. Please set na_action to the desired value to avoid seeing this warning
color_vector = pd.Categorical(values.map(color_map))
/opt/homebrew/Caskroom/mambaforge/base/envs/ascc24/lib/python3.9/site-packages/scanpy/plotting/_tools/scatterplots.py:394: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
cax = scatter(
/var/folders/md/d6lwwbv97xb6g6ddypntnprh0000gp/T/ipykernel_73405/1709870291.py:41: FutureWarning: The behavior of Series.replace (and DataFrame.replace) with CategoricalDtype is deprecated. In a future version, replace will only be used for cases that preserve the categories. To change the categories, use ser.cat.rename_categories instead.
query_map[query_coarse] = query_map[query_coarse].replace(to_replace=query_lut)
/var/folders/md/d6lwwbv97xb6g6ddypntnprh0000gp/T/ipykernel_73405/1709870291.py:42: FutureWarning: The behavior of Series.replace (and DataFrame.replace) with CategoricalDtype is deprecated. In a future version, replace will only be used for cases that preserve the categories. To change the categories, use ser.cat.rename_categories instead.
target_map[target_coarse] = target_map[target_coarse].replace(to_replace=target_lut)assert os.path.exists("hypl.json")
assert os.path.exists("hypl.html")You can visualize the plot in a notebook with:
# not run
plotly_overview.show()Pairwise comparisons
When comparing two clusters, it is most informative to have a look at the genes they are expressing. If the clusters are in the same species we can use a simple dotplot that combines the two clusters’ marker genes; things are not so simple when comparing between species. By plotting two dotplots side by side, and connecting homologous genes across the dotplots, we can get a better sense of how the clusters are connected.
A lot of the heavy lifting for this idea was already done by the scanpy team, who write excellent, well-documented code. I’ve adapted their code to produce the paired dotplots, fiddled with it to make sure dot size and color are consistent across plots, and added the homology lines. The latter part is rather tricky, and I would be thankful to anyone who can improve it; currently it involves a lot of heuristics to translate the coordinates from one side of the plot to the other.
paired_dotplot
paired_dotplot (query:anndata._core.anndata.AnnData, target:anndata._core.anndata.AnnData, connections:<built- infunctionarray>, query_clustering:str, target_clustering:str, query_species:str, target_species:str, query_cluster:Optional[str]=None, target_cluster:Optional[str]=None, pad:bool=True, x_offset:float=1, y_offset:float=0, grid_offset:float=30, query_gene_names:Optional[str]=None, target_gene_names:Optional[str]=None, output:str='./paired_dotplot.png', center:bool=True, title:Optional[str]=None, title_font_size:float=16, scale:bool=False, cmap:matplotlib.colors.Colormap='magma_r', layer:Optional[str]=None)
| Type | Default | Details | |
|---|---|---|---|
| query | AnnData | query species AnnData object | |
| target | AnnData | target species AnnData object | |
| connections | array | array of connected genes. Each row has at least two columns containing the query species gene and corresponding target species gene, and optionally their connection strength. Genes are allowed to be repeated on both sides. | |
| query_clustering | str | .obs column in the query AnnData object containing the query species clustering. |
|
| target_clustering | str | .obs column in the target AnnData object containing the target species clustering. |
|
| query_species | str | query species name. Will only be used in the title, so does not have to conform with the query species ID in the similarity matrix/SAMap object. | |
| target_species | str | target species name. Will only be used in the title, so does not have to conform with the target species ID in the similarity matrix/SAMap object. | |
| query_cluster | typing.Optional[str] | None | |
| target_cluster | typing.Optional[str] | None | the cell type/cluster of the query species that is being compared (default: None). |
| pad | bool | True | whether to pad the gene names with spaces to make them all of a similar length (default: True). |
| x_offset | float | 1 | Number of inches to add to the horizontal size of the canvas (default: 1). |
| y_offset | float | 0 | Number of inches to add to the vertical size of the canvas (default: 0). |
| grid_offset | float | 30 | Grid segments to add between the two dotplots. Might be useful if the gene names are not legible/lines overlap (default: 30). |
| query_gene_names | typing.Optional[str] | None | |
| target_gene_names | typing.Optional[str] | None | .var column that holds unique gene names for the query species (default: None). |
| output | str | ./paired_dotplot.png | path to save the plot to (default: “./paired_dotplot.png”). |
| center | bool | True | whether to center the dotplot (default: True). |
| title | typing.Optional[str] | None | overall title of the plot (default: None). |
| title_font_size | float | 16 | font size of the overall plot title (default: 16). |
| scale | bool | False | whether to scale the expression values to be between 0 and 1 for each gene (default: False). |
| cmap | Colormap | magma_r | colormap to use for the dotplot (default: “viridis”). |
| layer | typing.Optional[str] | None | layer : Union[str, None], optional The layer to use for the average expression calculation. If not specified, it will use the .X slot of the AnnData objects. It is vital to set this correctly to avoid calculating average expression on log1p-transformed data (default: None). |
| Returns | None |
Recall that scanpy dotplots need three inputs: the AnnData object, the names of genes to plot, and the name of the clustering. For paired dotplots we need this information for both sides of the plot.
However, genes are input via the connections slot; an array where each row contains two genes and (optionally) the strength of their connection. The function will reorder the genes in order to plot the densely connected gene groups first, and will also color-code each group to make reading the plot easier.
Additionally, since plots like this may become busy very fast, the function also accepts the names of the two clusters to be compared; it will then highlight the cluster names in the plot.
We are going to prepare the inputs first:
hydra = sm.sams["hy"].adata # the query dataset
planarian = sm.sams["pl"].adata # the target dataset
planarian.raw.var.index = "pl_" + planarian.raw.var.index
# the .obs column that contains the cluster labels
query_clustering = "Cluster"
target_clustering = "cluster"
# the species IDs
query_species = "hy"
target_species = "pl"
# the cluster labels to highlight
cluster_from = "ecEp_SC2"
cluster_to = "Epidermal: 2"The connections option is provided in this form deliberately, in order to allow the user to decide which genes to connect and how. This may cost a little extra effort for the most common cases, but it means that the function is more flexible.
The function currently supports three types of connections, corresponding to connection strength of [0, 1, 2], accordingly. It’s up to the user to assign meaning to the levels. The linestyles that correspond to these are
dottedfor 0 (linestyle = (0, (1, 5)))dashedfor 1 (linestyle = (0, (5, 5)))solidfor 2
In the future I would like to let the user specify the linestyles, but for now this is what we have.
Click here for some possible applications
- when comparing a dataset to itself, we could connect a gene to all its paralogs and visualize patterns of paralog substitution in cell type complement of an organism.
hydra_genes = np.array(
["hy_t13309aep", "hy_t14973aep", "hy_t10876aep", "hy_t38670aep", "hy_t19278aep"]
)
plan_genes = np.array(
[
"pl_dd_Smed_v4_361_0_1",
"pl_dd_Smed_v4_361_0_1",
"pl_dd_Smed_v4_1215_0_1",
"pl_dd_Smed_v4_1131_0_1",
"pl_dd_Smed_v4_1971_0_1",
]
)
orth_scores = np.array([0, 0, 1, 2, 2])
# connections without orthology scores
connections_plain = np.array([hydra_genes, plan_genes]).T
# connections with orthology scores
connections_orth = np.array([hydra_genes, plan_genes, orth_scores], dtype=object).TLet’s have a look at what we feed into the function:
pd.DataFrame(connections_plain)| 0 | 1 | |
|---|---|---|
| 0 | hy_t13309aep | pl_dd_Smed_v4_361_0_1 |
| 1 | hy_t14973aep | pl_dd_Smed_v4_361_0_1 |
| 2 | hy_t10876aep | pl_dd_Smed_v4_1215_0_1 |
| 3 | hy_t38670aep | pl_dd_Smed_v4_1131_0_1 |
| 4 | hy_t19278aep | pl_dd_Smed_v4_1971_0_1 |
paired_dotplot(
query=hydra,
target=planarian,
connections=connections_plain,
query_clustering="Cluster",
target_clustering="cluster",
query_species="hy",
target_species="pl",
query_cluster=cluster_from,
target_cluster=cluster_to,
pad=False,
)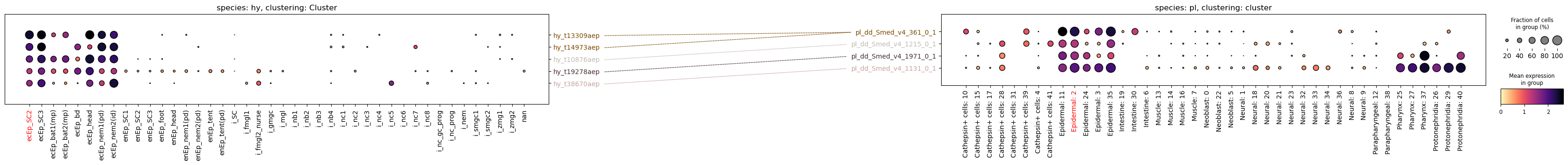
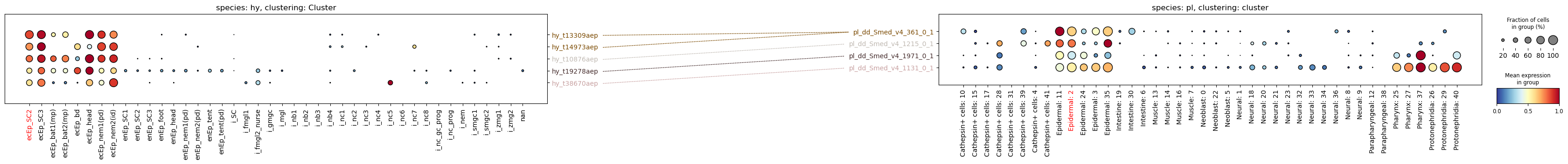
Gene expression can be very different between species, and plotting on the same scale can therefore be misleading. To address this, the function has a scale parameter that will normalize each row (gene) to a range of [0, 1]. This will make the plots more comparable, but it will also obscure the absolute expression levels. Use with caution.
Another thing to pay attention to: the scale function is not aware of log-transformed data, so if you applied log1p to your data it will calculate the an average-of-log instead of the log-of-average. This is a problem, and to avoid it please always use a layer that holds raw or normalised counts when using scale=True.
hydra.X.data = np.exp(hydra.X.data) - 1
hydra.layers["norm_counts"] = hydra.X.copy()
sc.pp.log1p(hydra)
planarian.X.data = np.exp(planarian.X.data) - 1
planarian.layers["norm_counts"] = planarian.X.copy()
sc.pp.log1p(planarian)paired_dotplot(
query=hydra,
target=planarian,
connections=connections_plain,
query_clustering="Cluster",
target_clustering="cluster",
query_species="hy",
target_species="pl",
query_cluster=cluster_from,
target_cluster=cluster_to,
pad=False,
scale=True,
cmap="RdYlBu_r",
layer="norm_counts",
)
Since we didn’t supply any weights for the connections, the function uses a weight of 0, and plots all connections we provided as dotted lines. Let’s try again, this time with weights:
WARNING - Gene name padding:
We used pad=False here because we used the gene IDs instead of gene symbols. Gene symbols tend to have the same length, and so pad doesn’t help here. In fact pad=True will throw an issue here, indicating possibly sloppy code. Unfortunately fixing this is not a priority.
pd.DataFrame(connections_orth)| 0 | 1 | 2 | |
|---|---|---|---|
| 0 | hy_t13309aep | pl_dd_Smed_v4_361_0_1 | 0 |
| 1 | hy_t14973aep | pl_dd_Smed_v4_361_0_1 | 0 |
| 2 | hy_t10876aep | pl_dd_Smed_v4_1215_0_1 | 1 |
| 3 | hy_t38670aep | pl_dd_Smed_v4_1131_0_1 | 2 |
| 4 | hy_t19278aep | pl_dd_Smed_v4_1971_0_1 | 2 |
paired_dotplot(
query=hydra,
target=planarian,
connections=connections_orth,
query_clustering="Cluster",
target_clustering="cluster",
query_species="hy",
target_species="pl",
query_cluster=cluster_from,
target_cluster=cluster_to,
pad=False,
scale=True,
cmap="RdYlBu_r",
layer="norm_counts",
)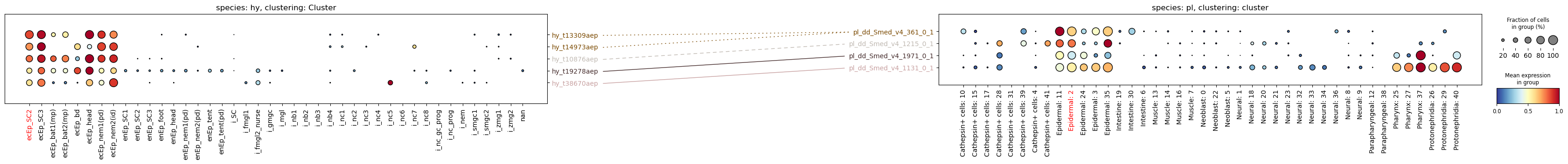
With weighting we can add an additional layer of information to the plot, such as orthology.
The next level would be to include the gene names, making this plot actually useful. For this to work reliably you need to have a column in the .var slot of your AnnData object that contains unique gene names. This is easily achieved by prepending the gene ID to the gene name/description.
The following code block is how I usually do this with EggNOG-mapper annotations; feel free to use whatever works for you.
Code
query = pd.read_csv(
os.environ["EXAMPLE_DATA_PATH"] + "eggnog/hydra.tsv", sep="\t", engine="python"
)
query = query[["Unnamed: 0", "5", "eggNOG_OGs", "21"]].copy()
query.columns = ["gene_id", "gene_name", "eggNOG_OGs", "description"]
query["gene_id"] = "hy_" + query["gene_id"].astype(str)
query.set_index("gene_id", inplace=True)
query["name"] = (
query.index.astype(str)
+ " | "
+ query["gene_name"].fillna("")
+ " | "
+ query["description"].fillna("")
)
hydra.var["name"] = hydra.var.index
hydra.var["name"] = hydra.var["name"].replace(query["name"].to_dict())
hy_t33417aep = "hy_t33417aep | EPT1 | diacylglycerol cholinephosphotransferase activity"
assert hydra.var.loc["hy_t33417aep"]["name"] == hy_t33417aepI have now created a column called name in the .var slot of the AnnData object. I can use this to plot the gene names instead of the gene IDs:
paired_dotplot(
query=hydra,
target=planarian,
connections=connections_orth,
query_clustering="Cluster",
target_clustering="cluster",
query_species="hy",
target_species="pl",
query_cluster=cluster_from,
target_cluster=cluster_to,
query_gene_names="name",
scale=True,
cmap="RdYlBu_r",
layer="norm_counts",
)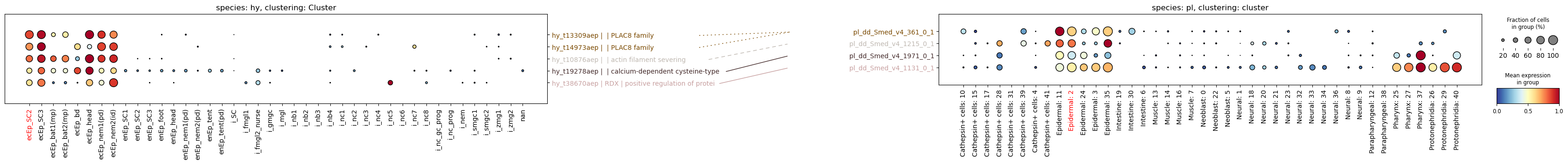
The right side of the plot is left as an exercise to the reader :) Notice how the lines on the left side are not starting from the same spot. This is where the util.procrustes function tries to guess how to pad using space characters so that the plotted strings have approximately the same length and the lines don’t go over some longer gene names. It’s not perfect, but it’s better than nothing.
From here on I will hide the lines of code that stay the same to make the notebook more readable. If you are trying to reproduce the plots, make sure you copy the code from the blocks above and just add the new lines.
The other options of the function should be self-explanatory, but I’ll go over them anyway:
x_offsetandy_offsetcontrol the canvas size beyond the size absolutely necessary for the plots to be rendered correctly. Increasex_offsetto give the plots more x-axis space, andy_offsetto give them more space on the vertical side. This is not an exact science, and you will find combinations that distort the plot to the point of making it unreadable.
paired_dotplot(
query=hydra, # |hide_line
target=planarian, # |hide_line
connections=connections_orth, # |hide_line
query_clustering="Cluster", # |hide_line
target_clustering="cluster", # |hide_line
query_species="hy", # |hide_line
target_species="pl", # |hide_line
query_cluster=cluster_from, # |hide_line
target_cluster=cluster_to, # |hide_line
query_gene_names="name", # |hide_line
x_offset=21,
y_offset=5,
scale=True,
cmap="RdYlBu_r",
)
The grid_offset parameter tries to control the space between the dotplots. Play with it if the lines connecting the genes are too short or too long. Try to stick to odd numbers, for some reason they work better. This gives you an additional lever for when increasing x_offset doesn’t work as intended.
I will also stop including the scale=True option, but will keep showing different color maps.
paired_dotplot(
query=hydra, # |hide_line
target=planarian, # |hide_line
connections=connections_orth, # |hide_line
query_clustering="Cluster", # |hide_line
target_clustering="cluster", # |hide_line
query_species="hy", # |hide_line
target_species="pl", # |hide_line
query_cluster=cluster_from, # |hide_line
target_cluster=cluster_to, # |hide_line
query_gene_names="name", # |hide_line
grid_offset=131,
x_offset=21,
cmap="viridis"
)
paired_dotplot(
query=hydra, # |hide_line
target=planarian, # |hide_line
connections=connections_orth, # |hide_line
query_clustering="Cluster", # |hide_line
target_clustering="cluster", # |hide_line
query_species="hy", # |hide_line
target_species="pl", # |hide_line
query_cluster=cluster_from, # |hide_line
target_cluster=cluster_to, # |hide_line
query_gene_names="name", # |hide_line
grid_offset=13,
x_offset=21,
cmap="cividis",
)
The parameters title and title_font_size do what you expect them to do.
paired_dotplot(
query=hydra, # |hide_line
target=planarian, # |hide_line
connections=connections_orth, # |hide_line
query_clustering="Cluster", # |hide_line
target_clustering="cluster", # |hide_line
query_species="hy", # |hide_line
target_species="pl", # |hide_line
query_cluster=cluster_from, # |hide_line
target_cluster=cluster_to, # |hide_line
query_gene_names="name", # |hide_line
x_offset=31,
y_offset=2,
title="If my title is long and in big font size I need to use the function\nparameters to make the plot bigger and push the plots to the sides",
title_font_size=40,
grid_offset=71,
cmap="plasma"
)
Finally, output controls where the plot is saved, and center=True will try to arrange the dotplots so that their centers on the y-axis are aligned. You probably want center=False if you need to print a title.
Real-life applications
When comparing cell types between species we are mostly interested in figuring out which cell types are homologous, and we want to find conserved gene expression patterns as evidence.
One way to get started with this is to look at the top marker genes of a cluster in one species and their homologs in the other animal.
For DEG detection I will use logistic regression, since I am interested in finding genes that are specifically expressed in my cluster of interest. I will take the "enEp_foot" cluster.
sc.tl.rank_genes_groups(hydra, "Cluster", method="logreg")
sc.tl.rank_genes_groups(planarian, "cluster", method="logreg")For the sake of illustration let’s have a look at the top 30 genes:
top30 = hydra.uns["rank_genes_groups"]["names"]["ecEp_SC3"][:30]sc.pl.dotplot(
hydra,
hydra.var["name"].loc[top30],
gene_symbols="name",
groupby="Cluster",
cmap="magma_r",
swap_axes=True,
use_raw=False,
)/opt/homebrew/Caskroom/mambaforge/base/envs/ascc24/lib/python3.9/site-packages/scanpy/plotting/_dotplot.py:168: FutureWarning:
The default of observed=False is deprecated and will be changed to True in a future version of pandas. Pass observed=False to retain current behavior or observed=True to adopt the future default and silence this warning.
/opt/homebrew/Caskroom/mambaforge/base/envs/ascc24/lib/python3.9/site-packages/scanpy/plotting/_dotplot.py:178: FutureWarning:
The default of observed=False is deprecated and will be changed to True in a future version of pandas. Pass observed=False to retain current behavior or observed=True to adopt the future default and silence this warning.
/opt/homebrew/Caskroom/mambaforge/base/envs/ascc24/lib/python3.9/site-packages/scanpy/plotting/_dotplot.py:747: UserWarning:
No data for colormapping provided via 'c'. Parameters 'cmap', 'norm' will be ignored
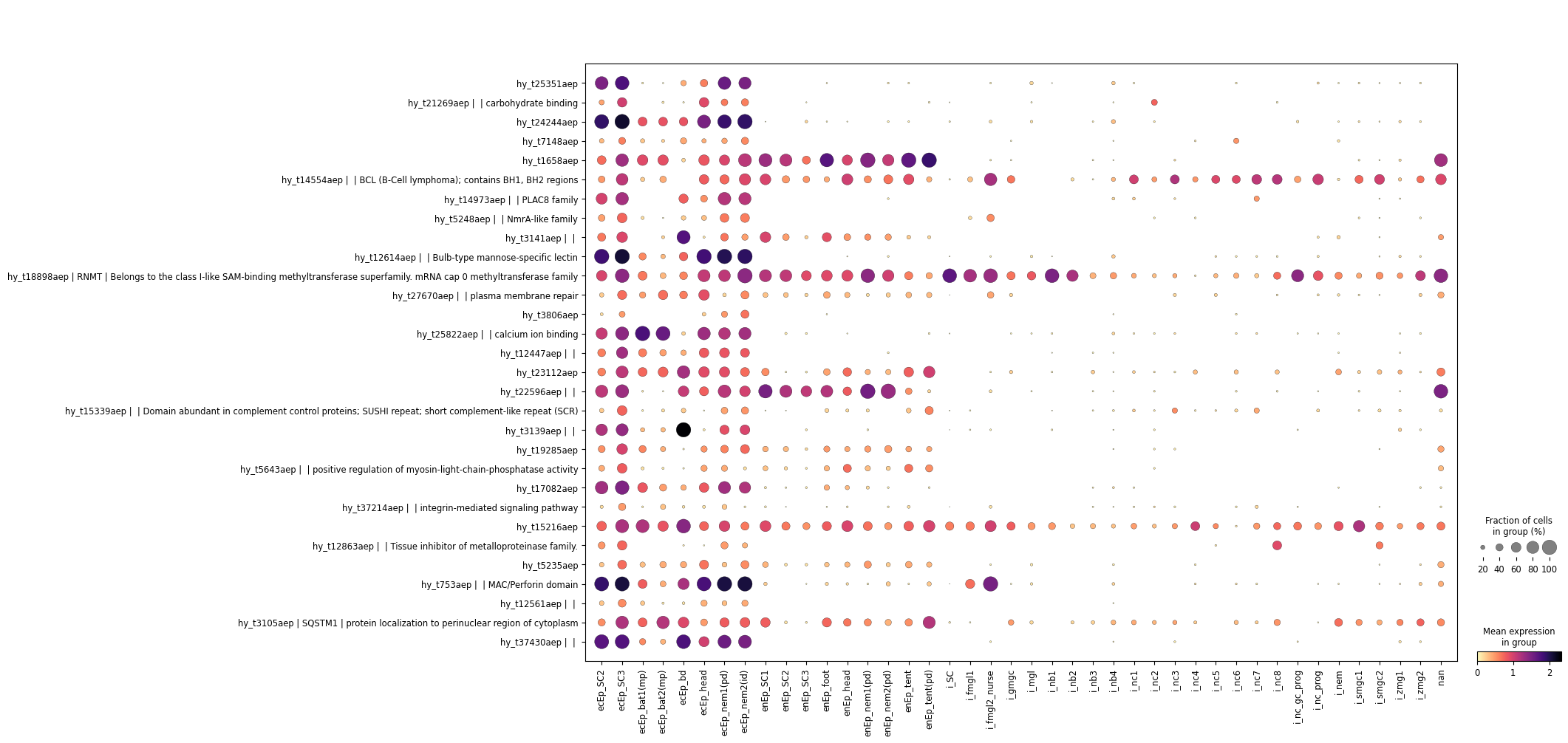
This might be too much information on the plot - many of the true marker genes are only expresed in the endodermal clusters anyway. I would like to collapse the other coarse clusters to make it more legible. I have a function for that:
util.collapse_unrelated_clusters(hydra, "ecEp_SC3", "Cluster", "coarse")sc.pl.dotplot(
hydra,
hydra.var["name"].loc[top30],
gene_symbols="name",
groupby="Cluster_collapsed",
cmap="magma_r",
swap_axes=True,
use_raw=False,
)/opt/homebrew/Caskroom/mambaforge/base/envs/ascc24/lib/python3.9/site-packages/scanpy/plotting/_dotplot.py:168: FutureWarning:
The default of observed=False is deprecated and will be changed to True in a future version of pandas. Pass observed=False to retain current behavior or observed=True to adopt the future default and silence this warning.
/opt/homebrew/Caskroom/mambaforge/base/envs/ascc24/lib/python3.9/site-packages/scanpy/plotting/_dotplot.py:178: FutureWarning:
The default of observed=False is deprecated and will be changed to True in a future version of pandas. Pass observed=False to retain current behavior or observed=True to adopt the future default and silence this warning.
/opt/homebrew/Caskroom/mambaforge/base/envs/ascc24/lib/python3.9/site-packages/scanpy/plotting/_dotplot.py:747: UserWarning:
No data for colormapping provided via 'c'. Parameters 'cmap', 'norm' will be ignored
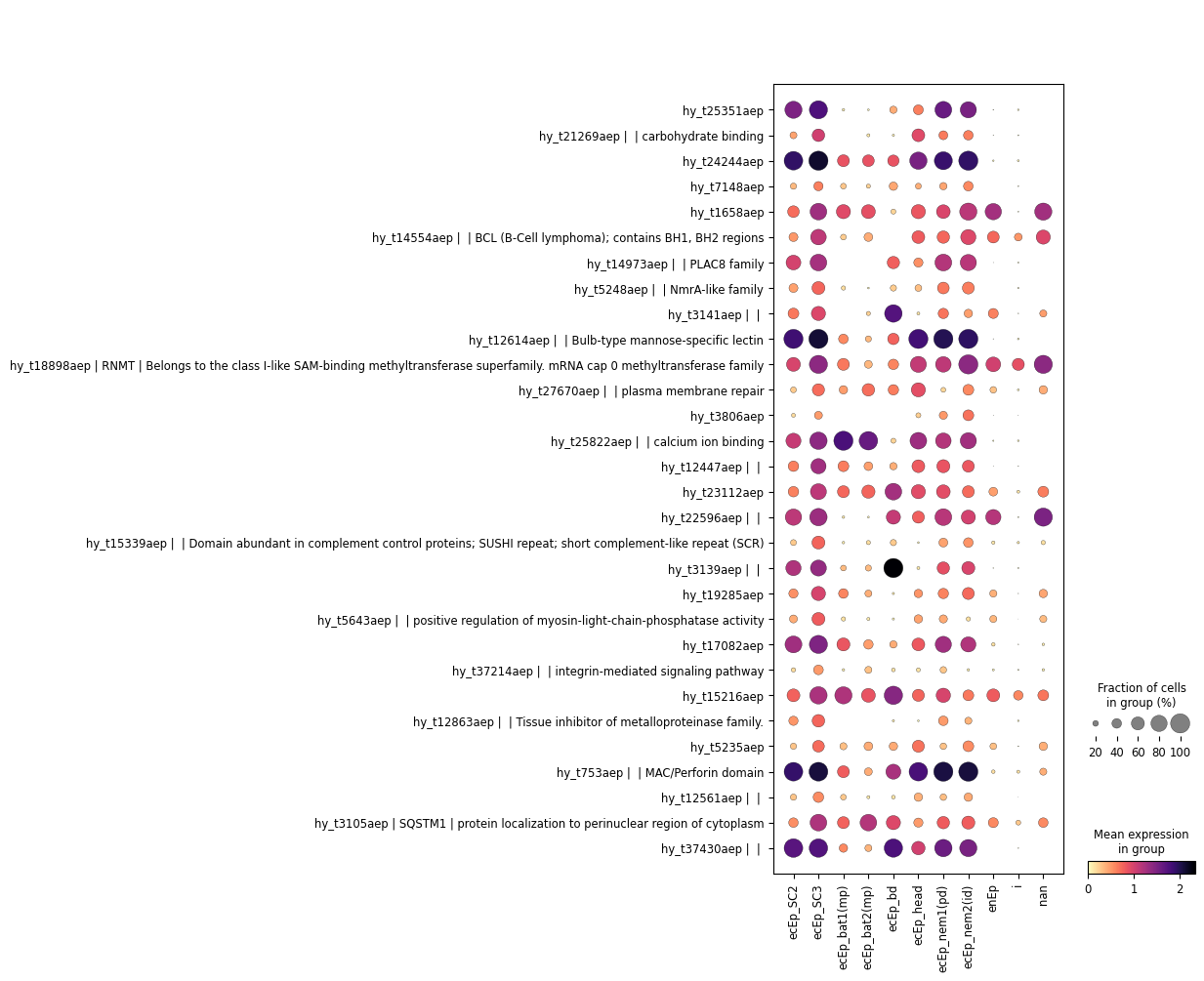
This is much more legible. Use this at your own risk though, as it may hide important information.
Now, let’s find the orthology information for these genes:
orthology = pd.read_csv(
os.environ["EXAMPLE_DATA_PATH"] + "hypl_orthology.tsv", sep="\t", index_col=0
)keep = np.intersect1d(
top30, orthology.index
) # for how many genes do we have orthology information?
available = orthology.loc[keep] # subset to only include those
available_long = available.reset_index().melt(
"gene_id"
) # pivot the table to long format
available_long| gene_id | variable | value | |
|---|---|---|---|
| 0 | hy_t12447aep | pl_dd_Smed_v4_10001_0_1 | 0 |
| 1 | hy_t12561aep | pl_dd_Smed_v4_10001_0_1 | 0 |
| 2 | hy_t12614aep | pl_dd_Smed_v4_10001_0_1 | 0 |
| 3 | hy_t12863aep | pl_dd_Smed_v4_10001_0_1 | 0 |
| 4 | hy_t14554aep | pl_dd_Smed_v4_10001_0_1 | 0 |
| ... | ... | ... | ... |
| 189970 | hy_t25822aep | pl_dd_Smed_v4_99_0_1 | 0 |
| 189971 | hy_t27670aep | pl_dd_Smed_v4_99_0_1 | 0 |
| 189972 | hy_t3105aep | pl_dd_Smed_v4_99_0_1 | 0 |
| 189973 | hy_t3139aep | pl_dd_Smed_v4_99_0_1 | 0 |
| 189974 | hy_t3141aep | pl_dd_Smed_v4_99_0_1 | 0 |
189975 rows × 3 columns
Now we can only keep the pairs with a score greater than zero:
connections = available_long[available_long["value"] > 0].to_numpy()We will now also name the planarian genes so that the plot makes more sense:
Code
target = pd.read_csv(
os.environ["EXAMPLE_DATA_PATH"] + "eggnog/planarian.tsv", sep="\t", engine="python"
)
target = target[["Unnamed: 0", "5", "eggNOG_OGs", "21"]].copy()
target.columns = ["gene_id", "gene_name", "eggNOG_OGs", "description"]
target["gene_id"] = "pl_" + target["gene_id"].astype(str)
target.set_index("gene_id", inplace=True)
target["name"] = (
target.index.astype(str)
+ " | "
+ target["gene_name"].fillna("")
+ " | "
+ target["description"].fillna("")
)
planarian.var["name"] = planarian.var.index
planarian.var["name"] = planarian.var["name"].replace(target["name"].to_dict())paired_dotplot(
query=hydra,
target=planarian,
connections=connections,
query_clustering="Cluster_collapsed",
target_clustering="cluster",
query_species="hy",
target_species="pl",
query_cluster="ecEp_SC3",
target_cluster="Epidermal: 2",
query_gene_names="name",
target_gene_names="name",
x_offset=15,
)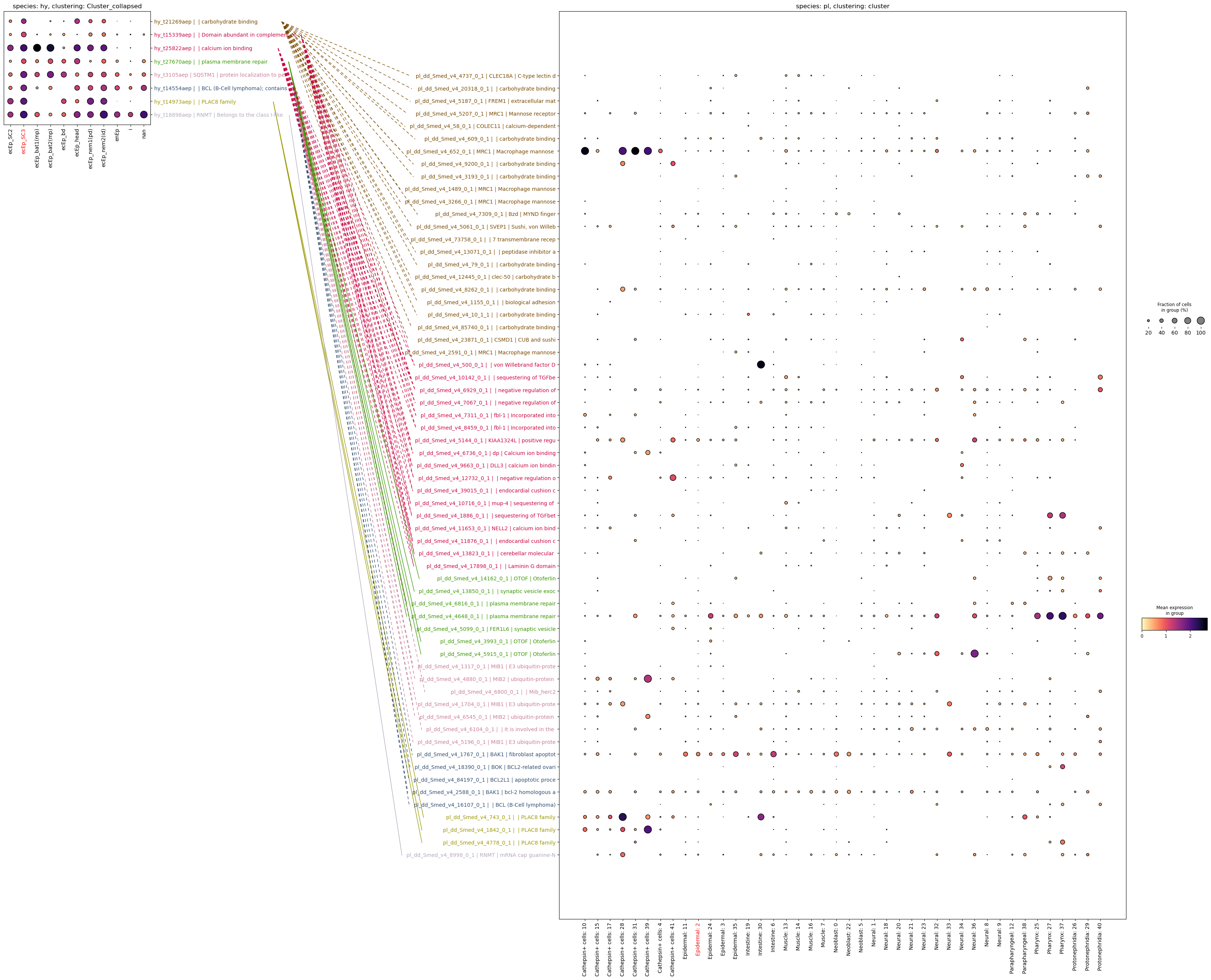
This approach will be sensitive but not very specific, as we will be plotting entire gene families on the target side. A more specific approach would be to look for genes that are markers on both sides and also homologous:
hydra_top100 = hydra.uns["rank_genes_groups"]["names"]["ecEp_SC3"][:100]
planarian_top100 = planarian.uns["rank_genes_groups"]["names"]["Epidermal: 2"][:100]connections = genes.get_orthologs_overlap(
hydra_top100, planarian_top100, hydra, planarian, orthology
)connectionsarray([['hy_t10711aep', 'pl_dd_Smed_v4_299_0_1', '1'],
['hy_t10711aep', 'pl_dd_Smed_v4_414_0_1', '1'],
['hy_t22760aep', 'pl_dd_Smed_v4_433_0_1', '1'],
['hy_t22760aep', 'pl_dd_Smed_v4_7131_0_1', '1'],
['hy_t22760aep', 'pl_dd_Smed_v4_762_0_1', '1']], dtype='<U22')paired_dotplot(
query=hydra,
target=planarian,
connections=connections,
query_clustering="Cluster",
target_clustering="cluster",
query_species="hy",
target_species="pl",
query_cluster="ecEp_SC3",
target_cluster="Epidermal: 2",
query_gene_names="name",
target_gene_names="name",
pad=False,
x_offset=-5,
)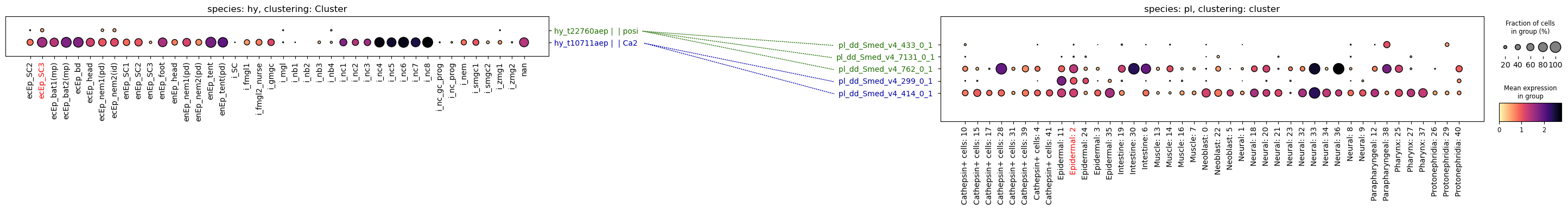
I clearly chose the wrong example here, but I hope you can see the idea behind the method, and how that could be useful when analysing your own data. Hopefully by comparing two species that are somewhat closer related than Hydra and Schmidtea.
Or, since the methods I describe here are so flexible, you could use them to manually compile lists of marker genes and visualize those, to much greater effect than this half-baked attempt at finding needles in a haystack. You probably know your favorite species much better than I know Hydra and Schmidtea, after all!
Or, and hear me out here, cnidarians might have a fundamentally different way of building their cell types compared to other metazoans, and so SAMap can’t find bonafide homologs. If we do all the pairwise comparisons and can’t find compelling evidence for homology, we can start to think about alternatives.
DISCLAIMER
Yes, I know that cnidarians and the rest of the metazoans had a common origin and that there is compelling evidence that many of their cell types are probably, in fact, homologous. I also know your in-situ data probably shows stuff that slightly disagrees with the single-cell data. I am just using hyperbole to make a point. Please don’t sue me.